First clssification of our clusters in the four main functional categories:
Knowns with PFAM (Ks): ORF clusters that have been annotated with a PFAM domains of known function.
Knowns without PFAMs (KWPs): clusters that have a known function, but do not contain PFAM annotations.
Genomic Unknowns (GUs): ORF clusters that have an unknown function (e.g. DUF, hypothetical protein) but are found in sequenced or draft-genomes.
Environmental Unknowns (EUs): ORF clusters of unknown function that are not found in sequenced or draft genomes, but only in environmental metagenomes.
Cluster of knowns
Pfam domain architectures in the clusters of knowns
Methods
We parsed the subset of annotated refined clusters to retrieve a single representative domain architecture for each cluster. The domain architecture is the linear order of the individual domains in multi-domain proteins. Most of the proteins in prokaryotes and eukaryotes are multi-domain proteins [1]. Studies of domain-based methods indicate that “comparing domain architecture is a useful method for classifying evolutionarily related proteins and detecting evolutionarily distant homologs” [2]. In case of clusters annotated to repeat (structural) motives, the representative domain architecture was defined using the suffix name of the domain followed by “_rep”, or “_n_rep” in case we have different families of the same repeat in the cluster. Within this step we divided the clusters annotated to Pfam Domains of Known Function (our set/category named knowns, or K) from those annotated to Pfam Domains of Unknown Function (DUFs), which became part of the GU category. A cluster has to be homogeneously annotated to DUFs (100% DUFs) to belong to the latter subset.
Results
Our entire dataset has 13,984 Pfam entries out of 16,172 (Pfam version 31). The 993,520 refined annotated clusters contain 9,871 different Pfam entries and are composed of 912,551 (92%) clusters annotated to Pfam domains of known function and 80,969 (8%) clusters annotated to Pfam domains of unknown function (DUFs). We retrieved a total of 29,864 original domain architectures (26,820 using reduced names, but keeping the repeats, and 23,781 using reduced names and contracting the repeats). From the whole set of clusters, we filtered out those annotated to DUFs that become part of the GU set.
The 912,551 clusters annotated to Pfam domain of known function contains 26,626 DAs (20,873 reduced ones), of which 13,321 (50%) contain more than one cluster, and consists of 780,452 clusters (96%) with mono-domain annotations and 132,100 with multi-domain annotations.
Cluster of unknowns
Methods
The subset of refined non annotated clusters was further processed to identify the two categories of unknowns: the genomic unknowns (GU) and the environmental unknowns (EU), and the second category of knowns: the knowns without Pfam (KWP). For this step we used a combination of searches against UniRef and NCBI nr protein databases.
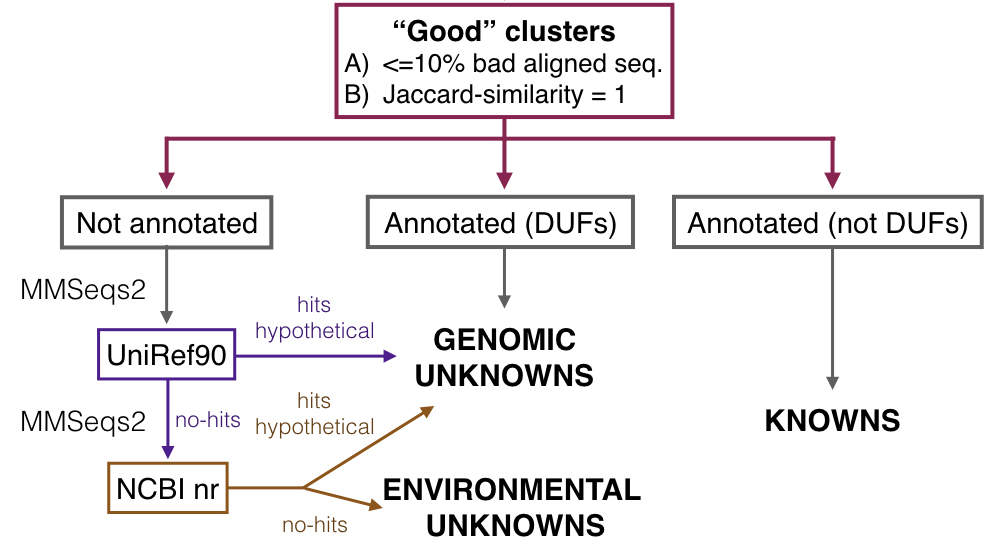
All the searches have the following characteristics:
1. Genomic unknowns: search against the Uniref90 database
In order to relate the filtered “good” subset of clusters of unknowns with the “hypothetical/uncharacterized” proteins found in sequenced genomes, we screened the not annotated cluster consensus sequences, retrieved from the MSAs of the compositional validation step, against the UniProt Reference Clusters (release 2017_11) from the UniProt Knowledgebase [3]. Specifically we searched UniRef90, which provides clusters of sequences with a sequence similarity > 90%. The search was performed using the search command of the MMSeqs2 software suite [4].
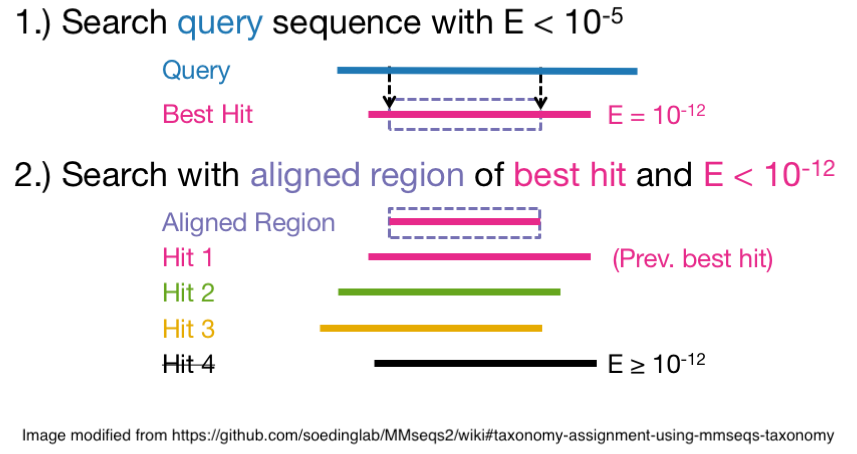
More specifically we performed a “double search”: 1) search of query sequences with E-value ≤1e-05 and query coverage ≥0.6; 2) extraction of the aligned region of the best hits and perform a second search; 3) merging of the top hits from the first search with the second search results; 4) filtering out the hits from the second search that have an E-value bigger than the first search top hits.
We classified each consensus sequences of the clusters as GU if all the hits that fall in the 60% of the best log10 e-value resulted annotated to a list of terms commonly used to define protein of unknown function in public databases, such as “hypothetical” and “uncharacterized” (“Hypothetical proteins” together with “uncharacterized protein” are the only recommended terms for naming proteins of unknown function (http://www.uniprot.org/docs/gennameprot). These terms imply that the structural genes do exist but the corresponding translation products have not been isolated and characterized yet.), Pfam Domains of Unknown Function (DUF), etc. The non-annotated clusters with a match to characterised proteins were considered (belonging to the) KWP class.
2. Environmental unknowns: search against the NCBI non-redundant database
The sequences that did not report any matches with UniRef90 entries were searched against the NCBI non-redundant (nr) database (release 2017_12) (http://www.ncbi.nlm.nih.gov) [5]. The NCBI nr is the single largest publicly available protein resource [6]. For the search we downloaded the fasta format file from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/nr.gz ) and used the MMseqs2 software [4] with the same parameters as described in the previous section. Those queries without any match to nr entries represent the EU set. The not annotated clusters matching nr entries with descriptions corresponding the list of terms mentioned above were considered GU, while those annotated to characterise entries KWP.
Results
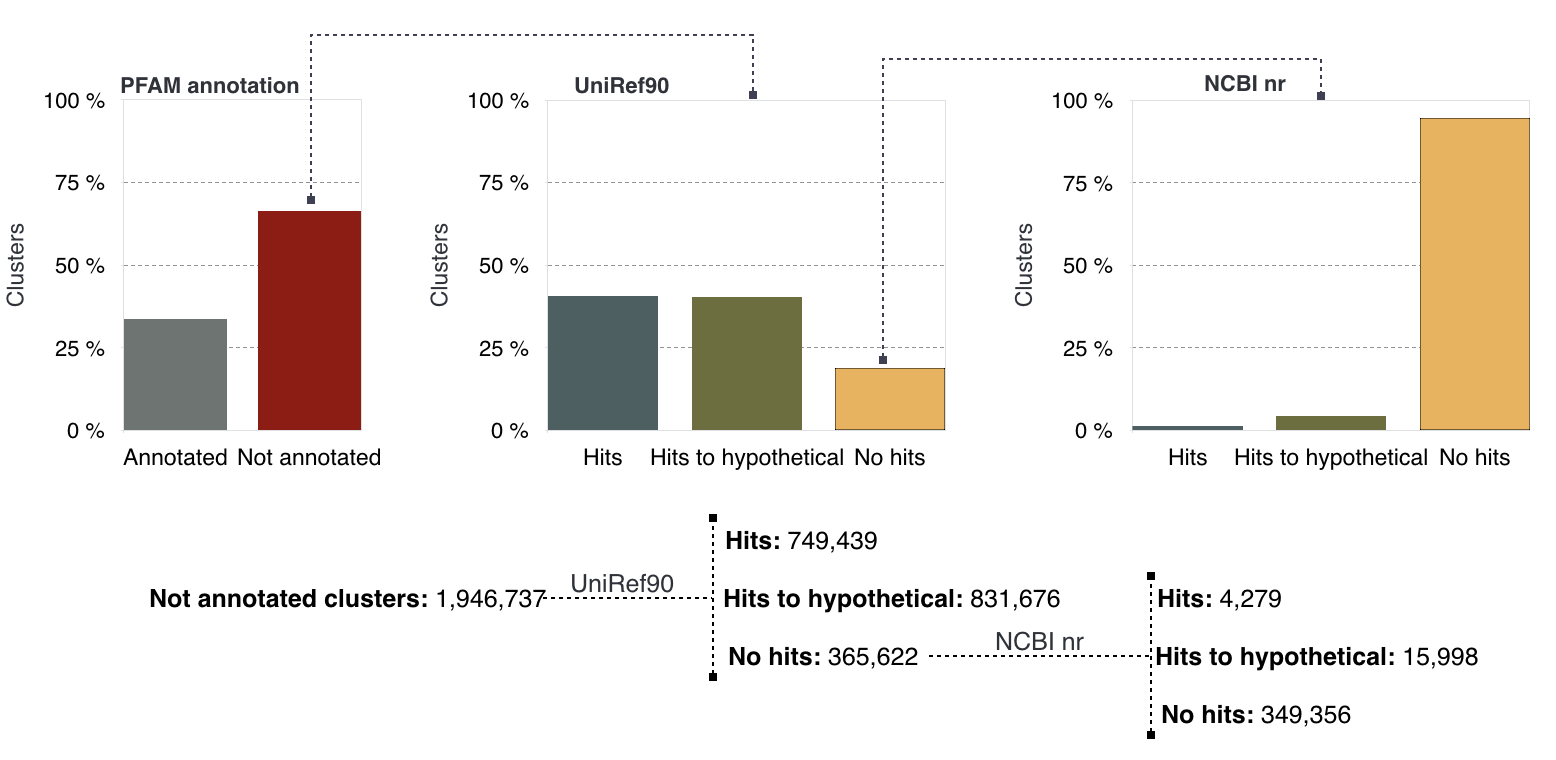
Of the 1,946,737 refined not annotated clusters, ∼1.5M reported a match (81%) to the UniRef90 database, and more than 50% of these hits resulted annotated to “hypothetical” proteins. The remaining ~400K (19%) clusters with no matches to UniRef90 entries were screened against the NCBI nr database. This second search resulted in ∼20K (5.5%) clusters with a match to NCBI nr entries, divided into ~16K clusters (79%), annotated to “hypothetical” proteins and ~4K (21%) clusters to not hypothetical. The remaining majority, ∼350K clusters (94.5%), reported no matches to NCBI nr entries. The results of the different steps are shown in the next figure, and the numbers reported in the table below.
Panel describing the steps applied to classify the cluster of unknowns
Results from the search against the UniRef90 and the NCBI nr databases
| Database | Characterised hits | Hypothetical hits | No-hits |
|---|---|---|---|
| UniRef90 | 749,439 | 831,676 | 365,622 |
| NCBI nr | 4,279 | 15,998 | 345,345 |
Pre-refinement/Preliminary cluster categories
We defined ORF clusters categories based on both the Pfam annotations and the results of the database searches. All the clusters annotated to a Pfam domain of known function represent the Knowns, and amount to ∼ 912K clusters (31%). The clusters annotated to DUFs, together with those annotated to hypothetical proteins constitute the genomic unknowns subset of ∼930K clusters (31%). The not annotated clusters without any match in UniRef90, nor in the NCBI nr database, amount to ∼350K (12%) and represent the environmental unknowns. The remaining ∼750K clusters (26%), not annotated to a Pfam domain, but reporting matches to UniRef90 or nr not-hypothetical proteins represented the KWP.
Results are summarised in the following table and figure:
Cluster categories and ORFs content.
| K | KWP | GU | EU | |
|---|---|---|---|---|
| Clusters | 912,551 | 753,718 | 928,643 | 345,345 |
| ORFs | 164,720,321 | 37,907,627 | 46,689,540 | 10,824,866 |
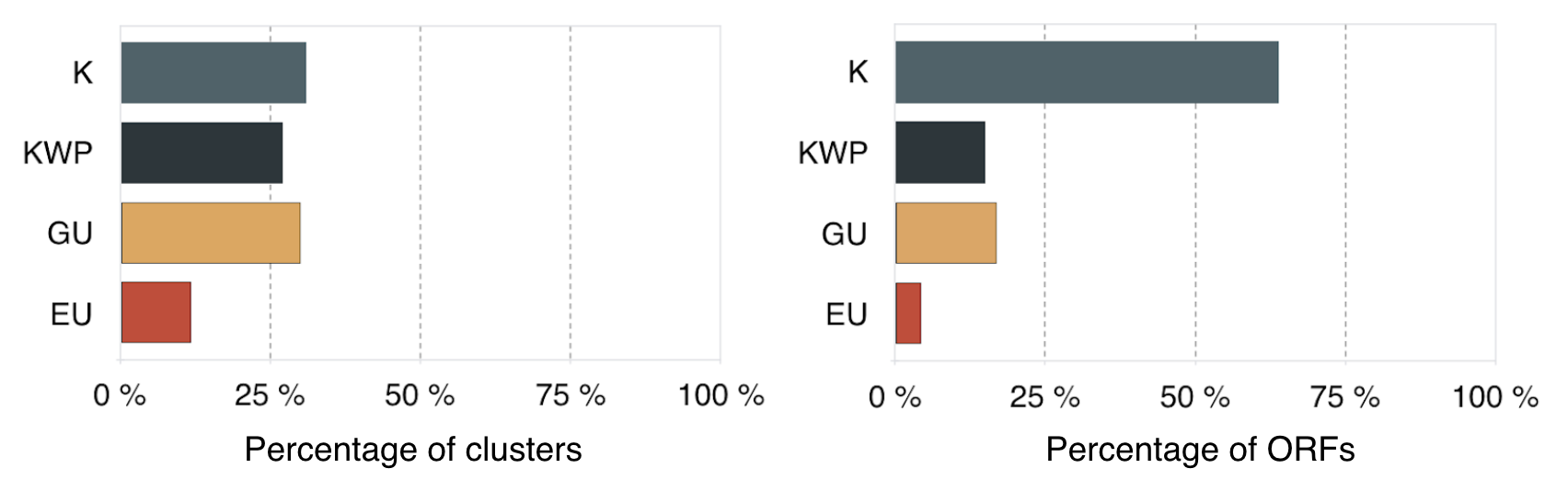
Percentage of clusters and ORFs in the different categories.
The main script, classification.sh, calls both the cluster of unknowns and the clusters of knowns classifications. The input are the refined set of annotated and not annoated clusters. Additional info in the README.
[1] H. Tordai, A. Nagy, K. Farkas, L. Bányai, and L. Patthy, “Modules, multidomain proteins and organismic complexity.,” The FEBS journal, vol. 272, no. 19, pp. 5064–5078, Oct. 2005.
[2] J. H. Fong, L. Y. Geer, A. R. Panchenko, and S. H. Bryant, “Modeling the evolution of protein domain architectures using maximum parsimony.,” J Mol Biol, vol. 366, pp. 307–315, Feb. 2007.
[3] M. Magrane and UniProt Consortium, “UniProt Knowledgebase: a hub of integrated protein data.,” Database: the journal of biological databases and curation, vol. 2011, p. bar009, Mar. 2011.
[4] M. Steinegger and J. Söding, “MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets.,” Nature biotechnology, vol. 35, no. 11, pp. 1026–1028, Nov. 2017.
[5] NCBI Resource Coordinators, “Database resources of the National Center for Biotechnology Information.,” Nucleic acids research, vol. 42, no. Database issue, pp. D7-17, Jan. 2014.
[6] S. Yooseph et al., “The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families,” PLoS biology, vol. 5, no. 3, p. 16, 2007.