Metagenomic data
To obtain a comprehensive view of the microbial communities in different environments we combined four major/primary marine metagenomic datasets, which cover all the ocean regions at various depths and the Human Microbiome Project dataset [1] The Global Ocean Sampling Expedition (GOS) [2], the Tara Oceans expedition (TARA) [3], Malaspina [Council SNRC (CSIC). Malaspina expedition. Available at: http://www.expedicionmalaspina.es/, 2010] and Ocean Sampling Day (OSD) [4], form together one of the most extensive public marine data sets. The data from GOS originated from 80 samples at 70 different sampling sites; the Malaspina data set comprises data from 116 samples, taken at 30 different stations; the TARA data covers 141 different locations for a total 242 samples and OSD data belongs to 146 metagenomic samples taken at 139 different stations. We added to this dataset 1,249 HMP metagenomes, coming from 5 main body sites (“gastrointestinal tract”, “oral”, “airways”, “urogenital tract” and “skin”) and 18 specific sites. The numbers are shown in Table 1.
Metagenomic data sets
| Data set | Samples | Sites |
|---|---|---|
| TARA | 242 | 141 |
| Malaspina | 116 | 30 |
| OSD | 145 | 139 |
| GOS | 80 | 70 |
| HMP | 1,249 | 18 |
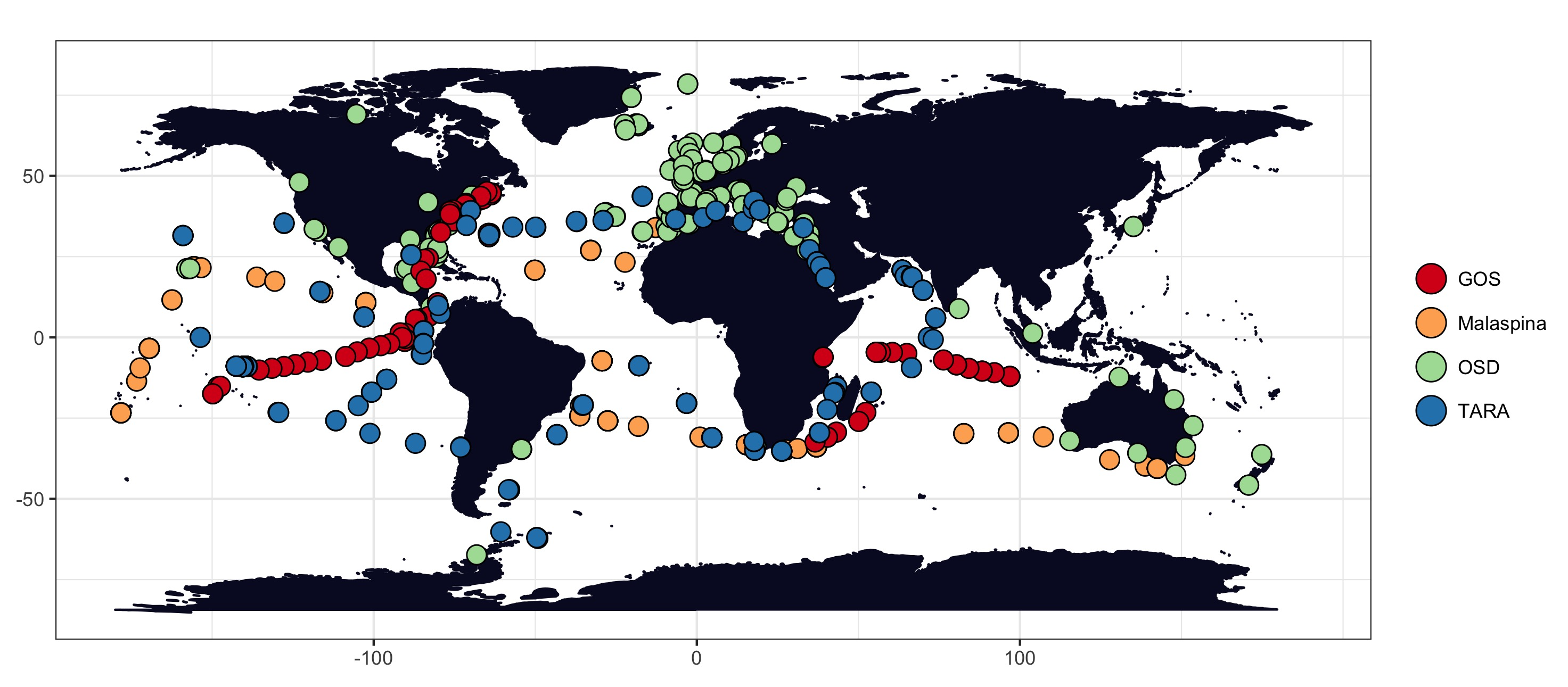
Ocean distribution of the metagenomic samples
The data were collected in the form of single-reads from GOS and at the stage of metagenomic assemblies from the other four projects. Specifically, GOS single-reads came from shotgun sequencing performed with the Sanger sequencing techniques, which leads to sufficiently long reads [5] (GOS Sanger data have an average read length of ~800 nucleotides [2]). TARA, OSD, Malaspina and the HMP data are, instead, metagenomic assemblies of Illumina pair-end reads. TARA reads were assembled using MOCAT [6], Malaspina with RAY-Meta [7], OSD using SPAdes [8] and the HMP with SOAPdenovo (V 1.04 28) [9].
Data sets integration via incremental clustering
Genomic data
The Genome Taxonomy Database (GTDB): 127,318 genomes, BACTERIA (125,243), ARCHAEA (2,075), Release 03-RS86 (19th August 2018)
We downloaded the protein sequences for bacterial and archaeal genomes from the Annotree website at: https://data.ace.uq.edu.au/public/misc_downloads/annotree/r86/.
We collected 90,621,864 proteins from 27,372 bacterial genomes, and 3,101,326 from 1,569 archaeal genomes
GTDB dataset
| Genomes | Proteins | |
|---|---|---|
| Bacterial | 27,372 | 90,621,864 |
| Archaeal | 1,569 | 3,101,326 |
| Total | 28,941 | 93,723,190 |
TARA gene catalog (version 2)
OM-RGC-v2 reference paper: “Gene Expression Changes and Community Turnover Differentially Shape the Global Ocean Metatranscriptome” https://www.sciencedirect.com/science/article/pii/S009286741931164X
OM-RGC.v2 contains 46,775,154 non-redundant genes.
It can be downloaded from the https://www.ocean-microbiome.org/ portal.
References
[1] J. Lloyd-Price et al., “Strains, functions and dynamics in the expanded Human Microbiome Project.,” Nature, vol. 550, no. 7674, pp. 61–66, Oct. 2017.
[2] D. B. Rusch et al., “The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific,” PLoS Biology, vol. no. 3, p. 77, 2007.
[3] S. Sunagawa et al., “Ocean plankton. Structure and function of the global ocean microbiome.,” Science (New York, N.Y.), vol. 348, no. 6237, p. 1261359, May 2015.
[4] A. Kopf et al., “The ocean sampling day consortium.,” GigaScience, vol. 4, p. 27, Jun. 2015.
[5] F. Sanger, S. Nicklen, and A. R. Coulson, “DNA sequencing with chain-terminating inhibitors.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 74, no. 12, pp. 5463–5467, Dec. 1977.
[6] J. R. Kultima et al., “MOCAT: a metagenomics assembly and gene prediction toolkit.,” PloS one, vol. 7, no. 10, p. e47656, Oct. 2012.
[7] S. Boisvert, F. Raymond, E. Godzaridis, F. Laviolette, and J. Corbeil, “Ray Meta: scalable de novo metagenome assembly and profiling,” Genome biology, vol. 13, no. 12, p. 122, 2012.
[8] A. Bankevich et al., “SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.,” Journal of computational biology: a journal of computational molecular cell biology, vol. 19, no. 5, pp. 455–477, May 2012.
[9] R. Li et al., “De novo assembly of human genomes with massively parallel short read sequencing.,” Genome research, vol. 20, no. 2, pp. 265–272, Feb. 2010.